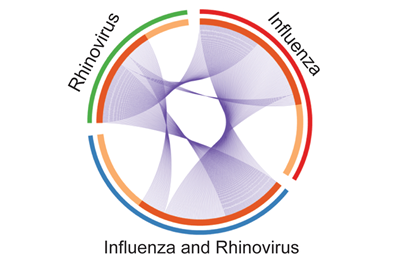
Gene changes caused by a given infection are marked as slices of the pie. Shared genomic changes between patients with influenza alone (red), human rhinovirus alone (green) or both viruses simultaneously (blue) are shown as ribbons connecting the slices of the pie.
Credit: Sarah Henrickson, MD, PhD, CHOP & Perelman School of Medicine
Acute respiratory tract infections (ARTI) are the leading global cause of death in early childhood, according to the Centers for Disease Control and Prevention (CDC). Lower respiratory tract infections, including bronchiolitis and viral and bacterial pneumonia, take a toll on children’s health, too, causing the majority of pediatric hospital admissions for infectious diseases.
By analyzing immune cells of children who came to the emergency department with flu symptoms, researchers found that the suite of genes these early-response cells expressed was shaped by factors such as age and previous exposures to viruses, according to a study by the Perelman School of Medicine at the University of Pennsylvania and Children’s Hospital of Philadelphia (CHOP). Better understanding how early infections influence long-term immune response has implications for the diagnosis and treatment of young patients who suffer from acute respiratory tract infections.
“The notion that an individual’s capacity to combat the flu depends on what they have been exposed to in the past, especially early in life, has been gaining momentum,” said senior author E. John Wherry, PhD, a professor of Microbiology and director of the Institute for Immunology at Penn. Wherry and Sarah E. Henrickson, MD, PhD, an instructor in the Allergy-Immunology division at CHOP, published their findings in Cell Reports this week.
“This study started during the 2009 H1N1 flu epidemic to find out how host responses change with different viral infections,” said lead author Henrickson, who began this work as a CHOP clinical fellow and postdoctoral fellow in Wherry’s lab. Previous studies elsewhere had investigated influenza responses broadly, but she wanted to focus on changes in CD8 T cells, key anti-viral cells in pediatric patients with influenza, and ultimately connect those changes to clinical outcomes, such as severity of infection, future asthma, fever, and return visits to a physician.
“Children generally have a less complex infectious history and less co-occurring conditions than adult patients,” she said. “As a result we can more easily assess the immune response to an acute infection and test how immune history shapes responses to the new infection.”
Sounding the Alarm
CD8 T cells prepare the body for fighting foreign viruses by altering their own gene expression after sensing the alarm signals raised by cells in the lungs in response to acute respiratory tract pathogens. In this study, the CD8 T cell gene expression in acutely ill pediatric patients with influenza-like illness was distinct from patients with other viral pathogens, such as rhinovirus. In general, the “genomic circuitry” of a cell – clusters of genes akin to electrical circuits that affect each other’s expression – varies according to the type of pathogen.
Using blood samples from 29 children who came to the CHOP emergency department with flu symptoms, the team found that different viruses elicit different immune responses – specifically, different patterns of genomic circuitry in CD8 T cells. Although these differences included the expected upregulation of interferon-stimulated genes and tamping down of cell adhesion proteins and signaling molecules, the pro-survival gene BCL2 was prominent in children presenting with an acute influenza infection.
The team found that differences in severity of ARTI, asthma, sex, and age also influence the immune response in an individual child. For example, younger children’s CD8 T cell gene circuitry was different from older children’s, which correlated to whether the younger child was exposed to the flu virus or not at an earlier age. Younger children with antibodies to a flu virus (evidence of previous exposure) had a gene expression pattern similar to the older children’s patterns. While this initial study is too small to correlate patterns such as the presence of older-child gene expression to clinical outcomes, the team plans to address this question in the future.
From the immune information they gathered, the team developed an Influenza Pediatric Signature (IPS) consisting of a small set of genes that consistently increased or decreased in expression in CD8 T cells from patients with an acute influenza infection. The IPS is able to distinguish acute influenza from ARTIs caused by other pathogens.
“Although this IPS is unlikely to replace clinical virological diagnosis anytime soon, the strength of the IPS score may reflect the severity of disease and provide helpful information post infection,” Wherry said. “It may help focus investigations on the key pathways in this population in the future.”
For example, the IPS helped identify an age-based difference in genome circuits related to the STAT1/2 pathway, which aids T cells to sense the inflammatory alarm raised by infected lung tissue and turn on interferon-stimulated genes to fight the virus. The IPS showed that the STAT1/2 circuit operates in young children with previous exposure to influenza (or the vaccine) similar to older children. This data suggests that therapies targeting the STAT1/2 pathway may be fruitful or that monitoring these signatures could be used to determine whether a vaccine works. The team hopes to investigate the importance of this altered circuitry in relation to clinical outcomes in larger studies going forward.
The researchers’ hope is that by combining the basic science of immune cell gene expression to actual cases seen in a high-volume pediatric ED will identify key pathways involved in host-pathogen interactions and help improve treatments for kids with severe flu symptoms.
Co-first author Sasikanth Manne, from Penn, and Kathleen E. Sullivan and Susan E. Coffin, both from CHOP, were also authors of the study.
This work was funded by the National Institutes of Health (AI112521; AI2010085 5T32HD043021-13; 5K12HD043245-15), the Pennsylvania Allergy Education Research Fund, and the Commonwealth of Pennsylvania.
Penn Medicine is one of the world’s leading academic medical centers, dedicated to the related missions of medical education, biomedical research, excellence in patient care, and community service. The organization consists of the University of Pennsylvania Health System and Penn’s Raymond and Ruth Perelman School of Medicine, founded in 1765 as the nation’s first medical school.
The Perelman School of Medicine is consistently among the nation's top recipients of funding from the National Institutes of Health, with $550 million awarded in the 2022 fiscal year. Home to a proud history of “firsts” in medicine, Penn Medicine teams have pioneered discoveries and innovations that have shaped modern medicine, including recent breakthroughs such as CAR T cell therapy for cancer and the mRNA technology used in COVID-19 vaccines.
The University of Pennsylvania Health System’s patient care facilities stretch from the Susquehanna River in Pennsylvania to the New Jersey shore. These include the Hospital of the University of Pennsylvania, Penn Presbyterian Medical Center, Chester County Hospital, Lancaster General Health, Penn Medicine Princeton Health, and Pennsylvania Hospital—the nation’s first hospital, founded in 1751. Additional facilities and enterprises include Good Shepherd Penn Partners, Penn Medicine at Home, Lancaster Behavioral Health Hospital, and Princeton House Behavioral Health, among others.
Penn Medicine is an $11.1 billion enterprise powered by more than 49,000 talented faculty and staff.